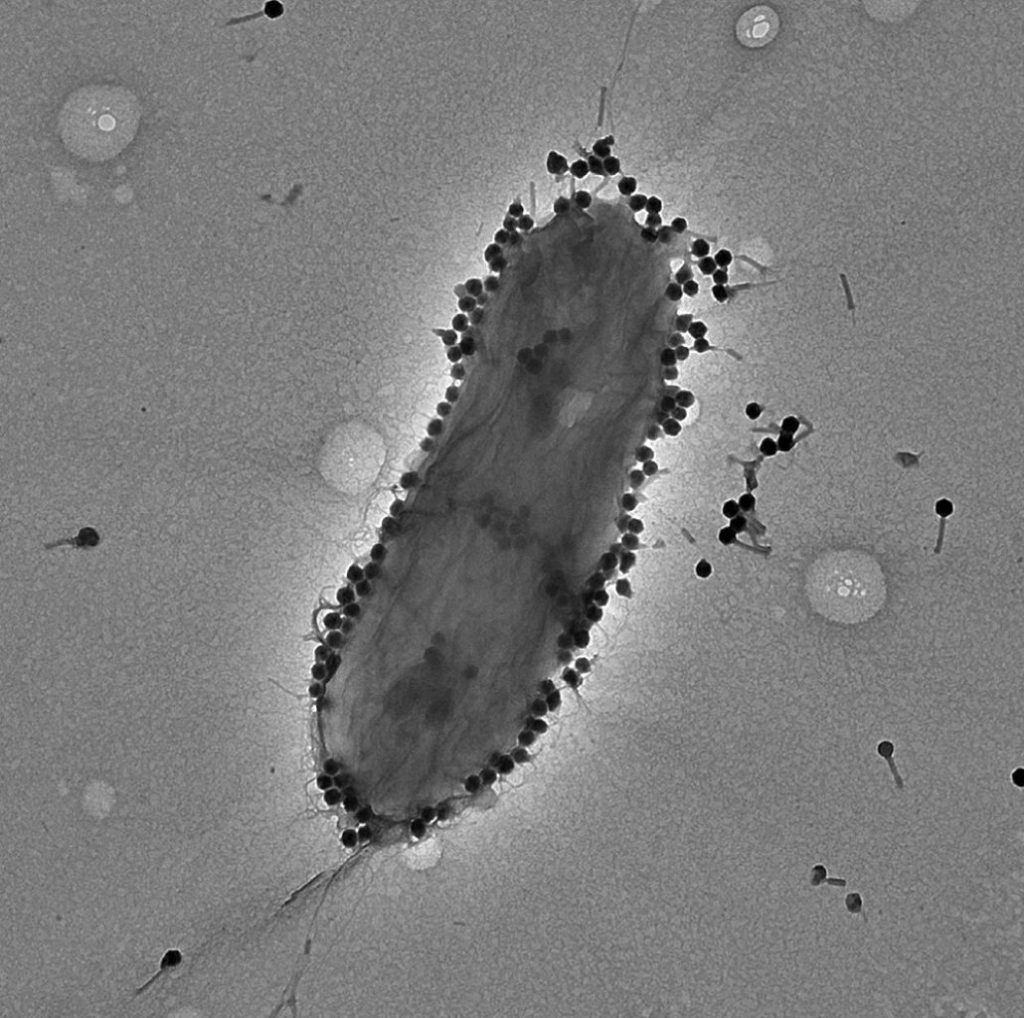
Many people are now familiar with phage therapy, a fascinating approach that harnesses viruses to combat drug-resistant bacteria. While this therapy boasts numerous benefits, there are some significant drawbacks that hinder its widespread adoption.
In contrast to antibiotics, which can be easily mass-produced in standardized production lines, phage therapy is often personalized for each case. This individualized approach poses challenges, as it can be time-consuming to prepare bacteriophages compared to the ready availability of antibiotics. To address this issue and enhance efficiency, scientists from Tulane University have successfully synthesized bacteriophages independently from bacterial cell cultures. This breakthrough offers a promising glimpse into a future where standardization can increase the efficiency in delivery of phage therapy.
Cell-free bacteriophage synthesis: How was it done?
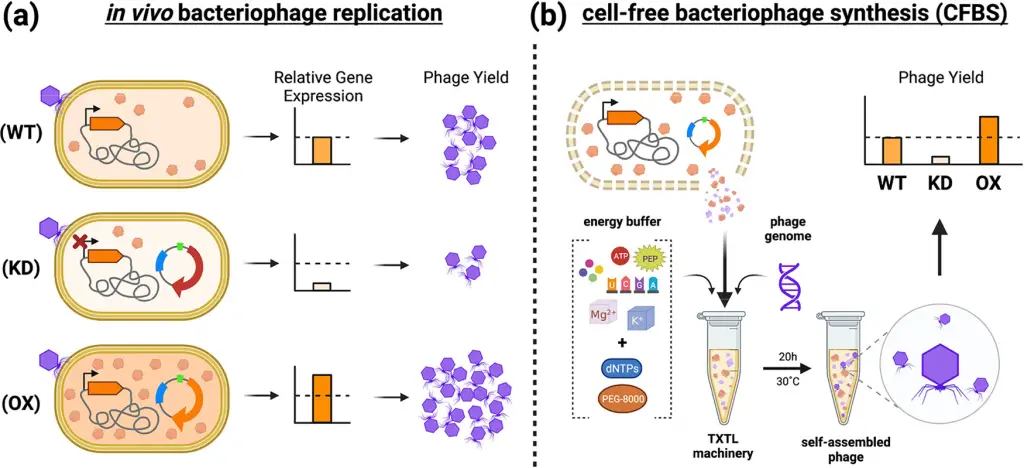
In a technique known as Cell-free Bacteriophage Synthesis (CFBS), scientists generated phage virions in a controlled environment outside of traditional laboratory settings that involve live bacteria. This innovative process replicates the natural in vivo phage production by utilizing cellular components extracted from bacterial lysates, such as transcription/translation machinery (TXTL), which is then combined with specific reagents to produce proteins based on the genetic instructions found in the DNA of the phage.
To put it simply, CFBS allows us to create phage virions without using live bacteria in a lab. It's like mimicking what happens inside a living cell but in a test tube, using cellular components and specific substances to make the proteins needed for the bacteriophage's replication when it infects a bacterial cell.
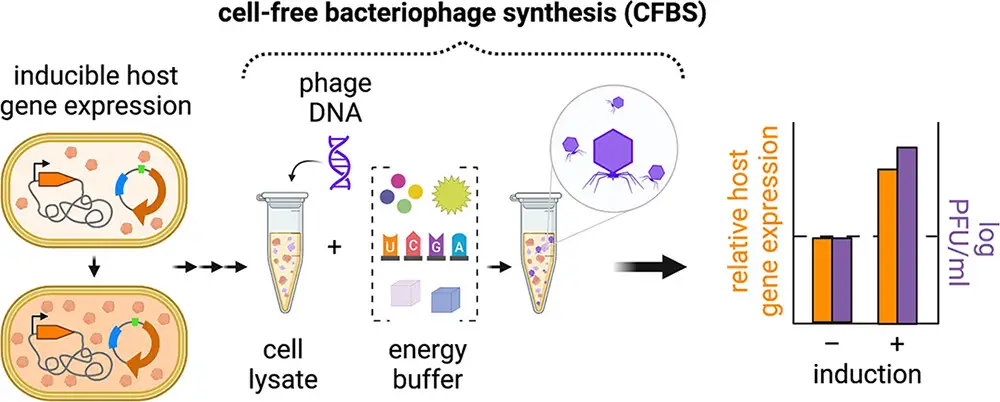
In this research, a process called TXTL (Transcription and Translation) was carried out using genetically modified E. coli BL21 to improve the production of the T7 bacteriophage in vitro through CFBS. To do this, they made alterations to the genetic makeup of these E. coli cells by manipulating 18 specific bacterial genes.
To manipulate these genes, the researchers employed a technique known as inducible CRISPR (read more about CRISPR Cas and other defense systems here) interference (CRISPRi), which involves an enzyme called Cas12a . This Cas12a enzyme, taken from a different bacterium (F. novicida), lacks nuclease activity, and it was used to identify the specific genes involved in T7 phage replication as either having a positive or negative impact.
Once the relevant genes were identified, the scientists further modified the TXTL's genetic background for CFBS. They did this by either increasing the activity of genes that had a positive effect (overexpressing) or reducing the activity of genes with a negative effect (repressing). This process allowed them to fine-tune the genetic environment within the TXTL to improve the production of the T7 bacteriophage in vitro.
Wonderful results
The results of this study were quite promising. They found that by making specific genetic modifications during the process of producing the T7 bacteriophage using CFBS, they were able to significantly increase the yield of these phages in vitro, sometimes by as much as 10 times.
The improvements were achieved through the following changes:
- Overexpression of the translation initiation factor IF-3 (infC).
- Enhanced levels of small RNAs OxyS and CyaR.
- Repression of the RecC subunit exonuclease RecBCD.
Translation initiation factors like IF-3 are proteins that play a crucial role in initiating the translation of genetic information into proteins, which is a fundamental step in building proteins. By manipulating these factors and small RNAs, the researchers optimized the process of creating T7 phages in the lab.
This is significant because it means that we can produce T7 bacteriophages more efficiently, potentially up to 10 times as effectively, by carefully adjusting the genetic components involved in their creation. By doing so, we can address some of the bottlenecks in phage manufacturing, making it easier and more practical to produce these viruses. This, in turn, can help lower barriers to the widespread use of phage therapy, which holds great promise as an alternative to traditional antibiotics in combating drug-resistant bacteria.
Reference and credits
Every image featured in this article originates from the same study that provided the information used in this article. To delve deeper into the details, please refer to the published study cited below. Brooks, R., Morici, L., & Sandoval, N. (2023). Cell Free Bacteriophage Synthesis from Engineered Strains Improves Yield. ACS Synthetic Biology, 12(8), 2418-2431.


Comments