Antibacterial resistance has led to the development of alternative treatments for bacterial infections, including the use of bacteriophages, which are viruses that can kill bacteria. Graham Hatfull, a professor of Biotechnology at the University of Pittsburgh, has been at the forefront of phage therapy, particularly for the treatment of chronic diseases such as cystic fibrosis. While phage therapy offers hope for treating bacterial infections, it is essential to understand how bacteria become resistant to phages, as resistance can limit the efficacy of this therapy.
Recently, Hatfull’s team discovered how a specific mutation in a bacterium, Mycobacterium smegmatis, results in phage resistance. The team isolated a mutant form of the bacterium that was resistant to infection by a phage called Fionnbharth and identified how the specific mutation in the lsr2 gene helped these resistant bacteria fight off a phage.
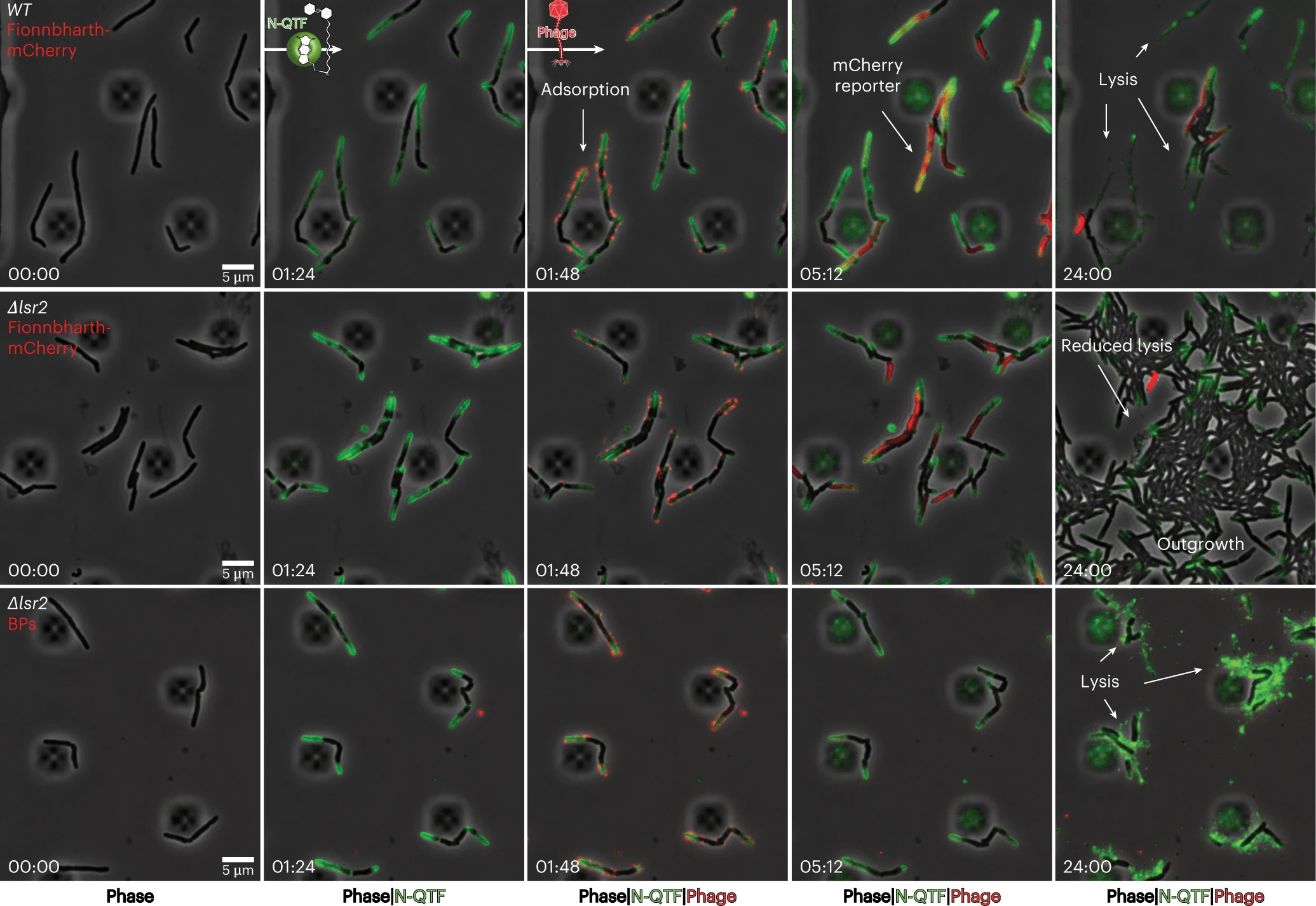
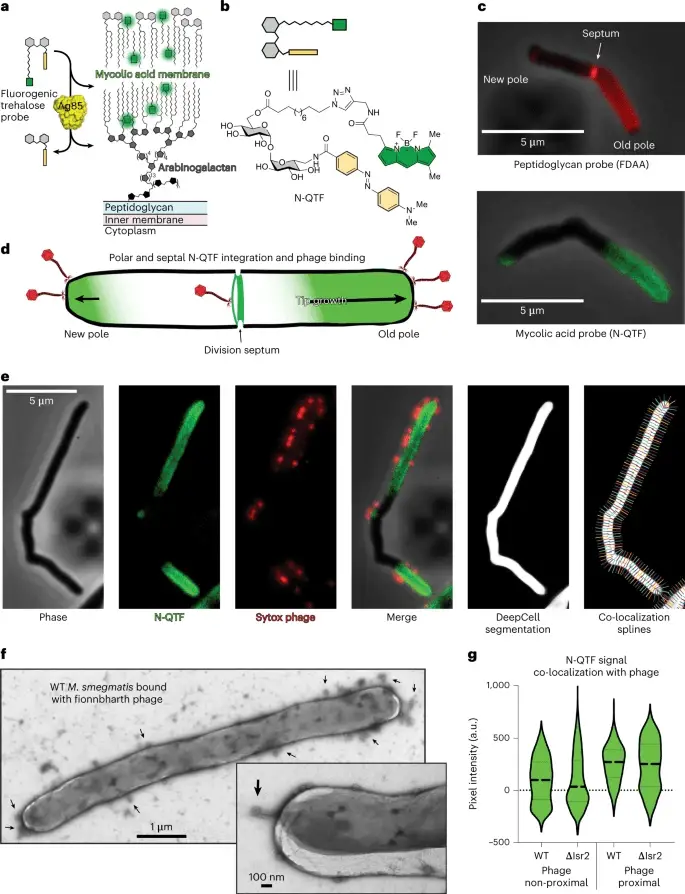
To understand how phages kill bacteria without the relevant mutation, Carlos Guerrero-Bustamante, a graduate student in Hatfull’s lab, genetically engineered two types of phages. One produced red fluorescence upon entering a bacterial cell aka reporter phage, while the other had segments of DNA that stuck to fluorescent molecules, allowing phage DNA to light up in an infected cell. This method enabled the team to watch, in unprecedented detail, as a phage attacked a bacterium.
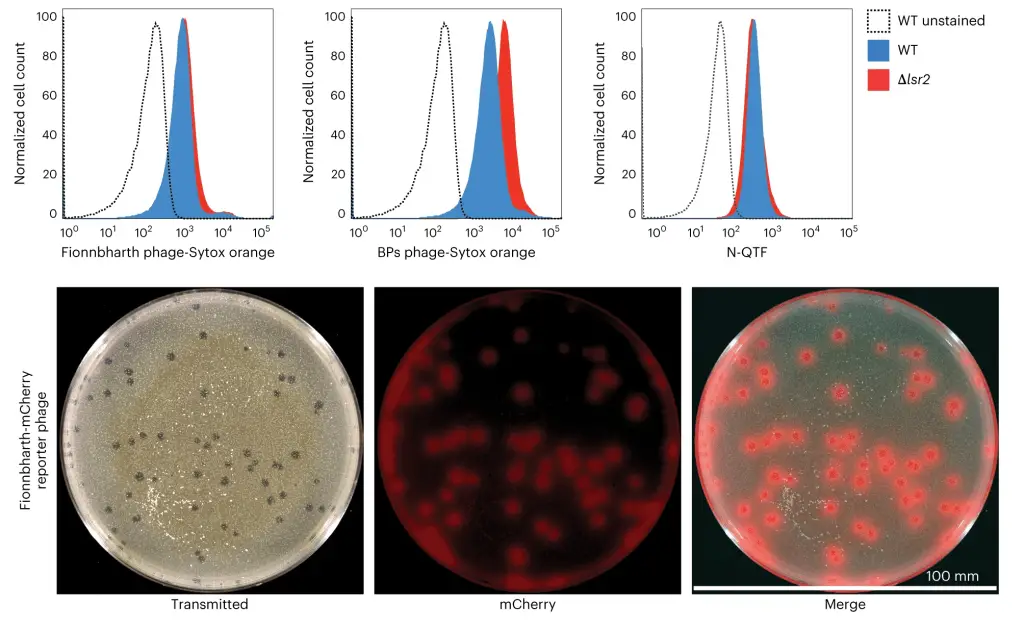
The team saw how phages bound to cells and injected their DNA into the bacteria. They then applied this knowledge to study the effect of removing the Lsr2 protein, which was critical in phage resistance. The link between Lsr2 and phage resistance was not previously known, but the team’s new methodology and tools made it clear that Lsr2 played a critical role in the replication of phage DNA.
Lsr2 typically helps bacteria replicate their own DNA. When a phage attacks, the virus co-opts the protein, using it to replicate phage DNA and overwhelm the bacteria. When the lsr2 gene is missing or defective, as in phage-resistant Mycobacterium smegmatis, the bacteria does not produce the protein, and phages do not replicate enough to take over the bacterial cell. The team’s discovery of the link between Lsr2 and phage resistance is a significant breakthrough that could help in the development of better phage therapies.
Hatfull emphasizes that this study focused on one bacterial protein and its resistance to just one phage. However, it has far-reaching implications, as there are numerous phages and proteins to study. The team’s new tools and methodology offer insights into how different mutations protect bacteria against invasion by phages. As phage therapy expands, these tools could help others better understand phages’ abilities while avoiding the missteps that led to antibiotic resistance.
The discovery of the link between Lsr2 and phage resistance is a significant step forward in understanding the mechanisms of bacterial resistance to phages. This knowledge could help in the development of more effective phage therapies to treat bacterial infections, including chronic diseases. It also highlights the importance of studying phages and their interactions with bacteria to identify new therapeutic options for antibiotic-resistant infections.
The new methodology and tools developed by Hatfull’s team offer an unprecedented level of detail into the interaction between phages and bacteria. This discovery of the link between Lsr2 and phage resistance is just one of the many possibilities that these tools offer. With the continued rise of antibiotic-resistant infections, phage therapy offers a promising alternative, and understanding bacterial resistance to phages is crucial in developing effective therapies.
This research has been published in a peer-reviewed journal and can be accessed via Charles L. Dulberger et al, Mycobacterial nucleoid-associated protein Lsr2 is required for productive mycobacteriophage infection, Nature Microbiology (2023). DOI: 10.1038/s41564-023-01333-x.
All photos used in this article are available on original published research work and all the credit goes to the researchers. Part of the conversation interview has been obtained from Phys.org