In the vast and intricate realm of microbiology, bacteriophages, known colloquially as phages, reign as some of the most prevalent and potent organisms on Earth. These viral predators specialize in targeting bacteria with lethal precision. Their existence and interactions with bacteria are fundamental to the dynamics of microbial ecosystems.
Recent research conducted by scientists at ADA Forsyth shed light on an aspect of this microbial battleground. Amidst the ongoing arms race between phages and bacteria, a third wheel has emerged—the ultrasmall bacterial parasites known as Saccharibacteria or TM7.
TM7 is an extremely small coccus (200-300 nm) bacterium with a unique parasitic lifestyle not previously observed in human-associated microbes. It is an obligate epibiont of various bacteria such as Actinomyces ssp., yet also undergoes a lethal phase, thereby killing its host. Their genomes are reduced, having less than 1MB without any amino acid biosynthetic capacity. An axenic culture of TM7 from the oral cavity was reported in 2014, but no sequence or culture was made available.
Published in the Proceedings of the National Academy of Sciences (PNAS), the study uncovered the role of this nanosized epibiotic parasite in conferring resistance to lytic phages upon its host bacterium, a strain of Schaalia odontolytica dubbed XH001.
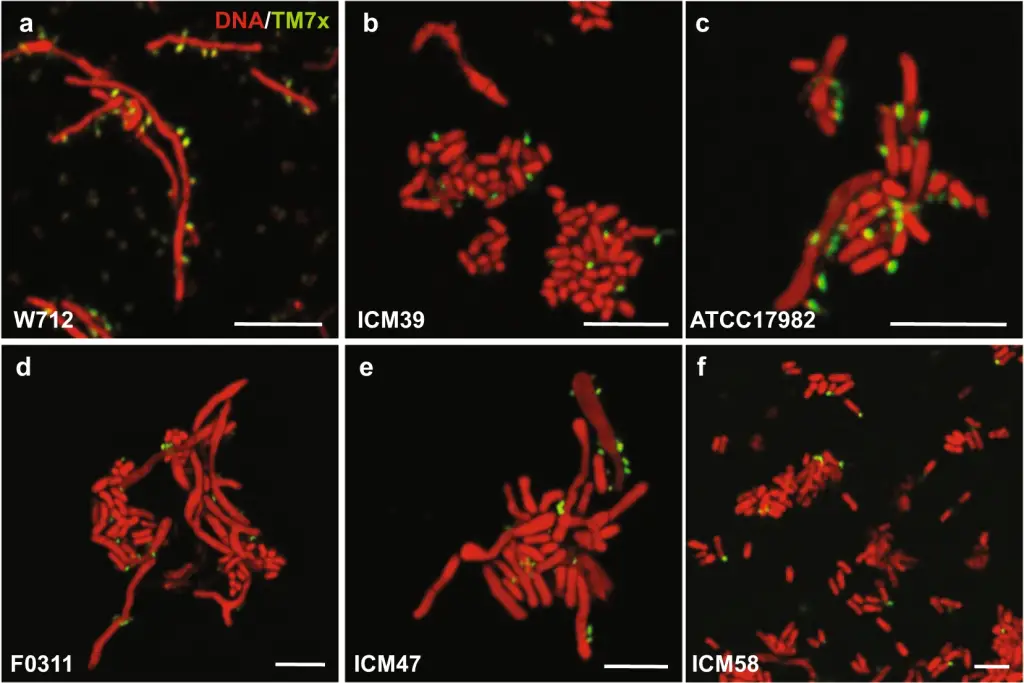
In laboratory conditions, TM7x exerts stress on the host bacterium, inhibiting its growth. However, within the complex microbial ecosystem of the oral cavity, TM7x assumes a beneficial role. By fostering a sink-source dynamic, TM7x facilitates the coexistence of phages and bacteria. Bacteria lacking TM7x remain susceptible to phage predation, while those harbouring the parasite resist phage attacks, thereby sustaining a balance between reproduction and defense.
TM7x exploits the host bacterium for its own benefit, it inadvertently aids the host in resisting phage infection—a novel interaction within the human microbiome. Unlike conventional phage-resistance mechanisms involving genetic mutations, TM7x induces physiological changes in the host bacterium, altering gene expression without altering its genetic makeup. This modulation of gene expression leads to surface structure modifications in the bacterium, rendering it impervious to phage attachment.
This groundbreaking research highlights the intricate relationships within microbial communities and underscores the importance of understanding the dynamics of phage-bacteria interactions for future therapeutic interventions and ecological management.
Have you liked what you have just read, click here to read more related content (phage-related news) like this one and also join our community by following the phage on X(formally Twitter), Facebook, and LinkedIn page
Study link: Le, Shuai et al, Episymbiotic Saccharibacteria TM7x modulates the susceptibility of its host bacteria to phage infection and promotes their coexistence, Proceedings of the National Academy of Sciences (2024). DOI: 10.1073/pnas.2319790121.