Integrated Phage Host Prediction, or iPHoP, is a command-line pipeline for automatically determining the host genus of new bacteriophages and archaea viruses based on those organisms’ genomic sequences. Imagine viruses as cunning toolkits, waiting for a chance to shine. But here’s the catch: these viruses are like picky guests at a party—they demand a specific host to operate. But, hold on, not just any host will do. It’s a particular host, one that the virus has evolved to master. Wait, did someone say hosts? Well, in this case, we’re talking about tiny microbes, such as bacteria, rather than humans. Now, with a touch of cutting-edge science known as metagenomic sequencing, we’ve stumbled upon more of these viruses than we ever dreamed of, spanning diverse ecosystems. Yet, there’s a puzzle to solve: matching these virus genes to their microbial partners is like cracking a secret code that helps us grasp the virus’s capabilities. And that’s where the star of our show steps in—the iPHoP program (pronounced “eye-pop”), a dazzling new innovation in the world of viruses and their microscopic companions.
The Thrill of the Discovery
Picture this: our world is buzzing with trillions of microbes, the tiny rulers of their domains—archaea and bacteria. They call the shots when it comes to essential tasks like carbon recycling, nitrogen balancing, and even methane production. But guess what? These unseen environmental heroes are under constant viral assault, their scripts rewritten by sneaky viruses. Now, here’s where things get intriguing. If we can decipher the dance between viruses and their microbial counterparts, it’s like holding the key to a magical kingdom. Imagine using these viral matchmakers to manipulate microbial communities, possibly helping plants interact better, turbocharging nutrient cycles, or even predicting the best candidate for phage therapy.
Cracking the Viral Code with iPHoP
So, how do we crack the viral code? It’s like piecing together a cosmic puzzle, and the iPHoP program is our genius guide. You see, there are other programs out there trying to pair viruses with potential hosts, but they sometimes end up with different answers. iPHoP is like a master chef who brings together multiple recipes into one delightful dish and then adds a sprinkle of machine-learning magic. The result? A prediction that’s not only fused from various sources but also stamped with a confidence score, like a seal of approval. Imagine this prediction as a “genus-level host prediction tool,” a treasure map guiding us through the labyrinth of virus-host connections.
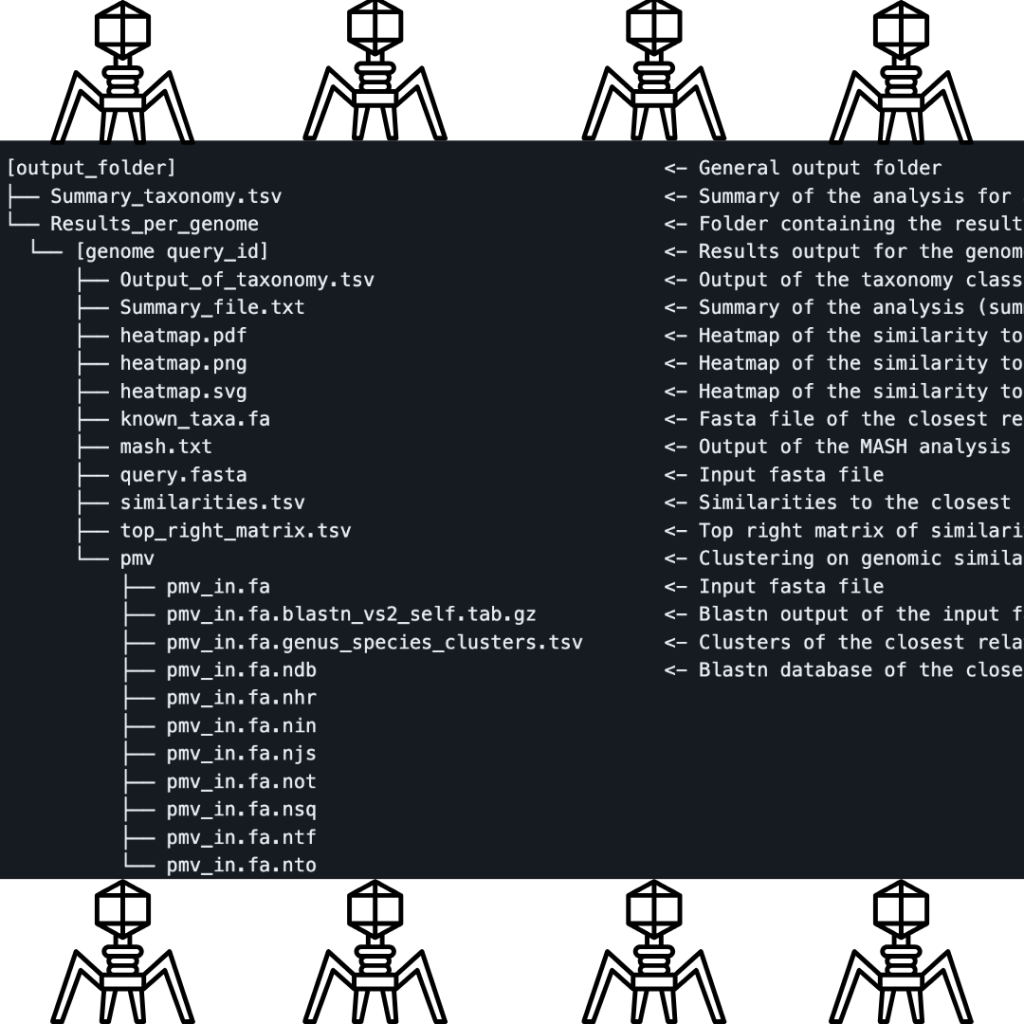
In a groundbreaking stride towards comprehending the intricate interplay between viruses and their microbial hosts, the iPHoP (Integrated Phage Host Prediction) pipeline stands as a game-changer. This computational marvel operates through a meticulously orchestrated sequence of six main steps, each unraveling a layer of the enigmatic puzzle.
Step 1: Unveiling Host Predictions
The journey begins with individual host prediction tools stepping into the spotlight. Picture a phage-based tool, RaFAH, yielding host genus predictions alongside associated scores, reserved for later assessment. Host-based tools join the fray with their unique contributions. “blastn” explores host genomes, isolating hits with identity scores ≥ 80% and lengths ≥ 500bp. However, hits spanning over 50% of the “host” contig length are dismissed, as they often result from nearly purely viral sequences, potentially introducing contamination. A twist emerges with “blastn” targeting CRISPR spacer databases, embracing hits with up to 4 mismatches. Meanwhile, tools like WIsH, VHM – s2* similarity, and PHP factor in k-mer composition similarities between virus and host genomes to predict associations.
Step 2: Bridging Scores and Distances
The second stride involves the meticulous assembly of scores and distances derived from host-based tool hits. The distances between potential hosts are carved out based on the GTDB trees, weaving an intricate web of connections.
Steps 3 and 4: Curating the Clues
Here, the pipeline delves into compiling well-organized lists of hits for each combination of virus, tool, and potential host. These hits are like breadcrumbs leading to automated classifiers, each weaving a unique score for the virus-host pairing. This intricate dance considers not only the base hit host but also the contextual relationships with other hits.
Step 5: Trifecta of Scores
A trio of scores emerges for host-based tools in the fifth phase. While retaining top scores solely based on “blast” or CRISPR matches, a third score blossoms, integrating the cumulative output from all individual classifiers. This cohesive strategy acknowledges the strengths of various host-based methods.
Step 6: Forging the Composite Score
The sixth and final step is akin to an alchemical fusion, where host-based and phage-based signals converge. The three host-based scores, carefully crafted in the previous steps, harmonize with the phage-based RaFAH score, birthing a singular score that encapsulates the essence of each virus-candidate host genus pair.
Databases and Versions: Behind the Scenes
iPHoP operates within specific databases and versions. The current iteration, v1.3.0, introduces a recoded WIsH module, necessitating adjustments to the host database structure. To align with this version, the “Sept_21_pub_rw” database is recommended by , incorporating GTDB release 202 genomes, public IMG genomes, and metagenome-assembled genomes from the GEM dataset. A “Test_db_rw” database caters to testing purposes, featuring a compact selection of host genomes tailored for the “test_input_phages.fna” file’s phage genomes.
As we journey deeper into the realm of viruses and their elusive host partners, the iPHoP pipeline emerges as a guiding beacon, weaving together predictive tools, scores, and relationships. Its intricate dance promises to unlock the mysteries of these microbial connections, forever reshaping our understanding of the microbial world.
The Birth of iPHoP: A Journey to Unravel Mysteries
Now, let’s rewind to the creation of iPHoP, a tale spun by the brilliant minds at the U.S. Department of Energy Joint Genome Institute. These scientific pioneers embarked on a mission—to streamline a process that often felt like choosing between multiple paths in a maze. They harnessed the power of a machine-learning model and set it loose on 1,870 known virus-host pairs. The outcome? A matrix of data points that fueled the machine’s intelligence. Then, like a detective piecing together evidence, they unleashed iPHoP on a whopping 216,015 top-notch virus sequences from the IMG/VR database. Hold onto your seats, because iPHoP unveiled a treasure trove of new host predictions across various landscapes. Imagine it as an explorer uncovering hidden secrets in human microbiomes or deciphering the tales viruses whisper to soil and crops.
Funding the Dream
Remember, dreams don’t come true without a little support. The U.S. Department of Energy, through its Biological and Environmental Research Wing, played fairy godparent. They granted funds under the Early Career Research Program, paving the way for these discoveries. The U.S. Department of Energy Joint Genome Institute, a powerhouse under DOE’s Office of Science, also played a crucial role through their unwavering support.
The Grand Finale: Opening Doors to New Possibilities
As the pages turn on this thrilling tale, remember that every discovery opens doors to possibilities. With iPHoP, we’re tapping into the hidden symphony between viruses and their microbe companions. It’s like unlocking the potential of our planet’s natural processes, from boosting crop yields to uncovering the roles of viruses and microbes in our environment.
Intrigued? Dive into the Details
For the curious minds hungry for more, you can explore the in-depth findings in the “PloS Biology” journal (Roux, S. et al. “iPHoP: An integrated machine learning framework to maximize host prediction for metagenome-derived viruses of archaea and bacteria.” PloS Biology 21(4): e3002083, 2023).
So, there you have it—the saga of iPHoP, the ultimate matchmaker that reveals the dance between viruses and their microbial hosts. Who knew that the tiniest players could have the greatest impact? And remember, the next time you think about viruses, consider them not just as threats, but as the architects of a microbial symphony that shapes our world.
We are eager to include this tool in our collection. If you’re aware of any other valuable phage-related tools that deserve a spot on our page, kindly let us know. Your input is highly appreciated.