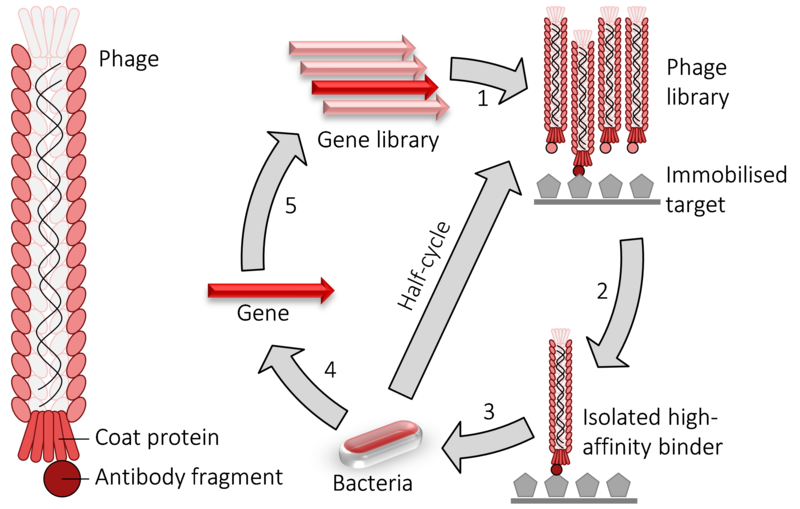
Phage display is a molecular biology technique where bacteriophages, viruses that infect bacteria, are genetically modified to express foreign proteins on their surface. This modification typically involves inserting a gene encoding the protein of interest into the genome of the bacteriophage in a region responsible for encoding the coat proteins of the phage. As a result, when the bacteriophage replicates, the foreign protein is displayed on the surface of the viral particles (virions). This allows for the presentation of a diverse array of proteins or peptides, which can then be screened for interactions with other molecules, such as antibodies or receptors, facilitating the study of protein-protein interactions, epitope mapping, and drug discovery, among other applications.
In this technique, in vitro selection entails the screening and amplification of extensive protein libraries, mirroring the principles of natural selection. Phage display stands out as a potent method for generating significant quantities of peptides, proteins, and antibodies. Among the filamentous bacteriophages commonly employed in phage display—namely, f1, fd, and M13—E. coli serves as the host organism. Nevertheless, researchers have also leveraged other bacteriophages like T4, T7, and phage, broadening the scope and applicability of this versatile approach.
In 1985, G. Smith pioneered this technique as a method for displaying polypeptides on the surface of lysogenic filamentous bacteriophages.
Applications of Phage Display
Phage display, as a fast-expanding technology, is now used in a broad range of research areas, such as epitope mapping, the identification of new receptors and ligands, in vitro protein evolution, drug discovery, and so on. The following sections explain some of the most successful phage display applications.
Epitope mapping and mimicking
When exposed to the antigen, the host’s humoral immunity is activated, resulting in the production of antibodies directed against foreign protein epitopes. Understanding the pathogenesis of pathogen infections and developing diagnostic reagents, therapeutic antibodies, and effective vaccines all rely on knowledge of these protein epitopes.
Peptide phage display libraries are useful for identifying continuous or linear epitopes that interact with antibodies. Mimotope isolation and identification from peptide phage libraries is a powerful approach for improving immunological studies in order to design and develop vaccine candidates.
Identification of new receptors & ligands
The phage display method, which uses either gene-specific libraries or random peptide libraries, is a powerful technique for epitope identification. The method can detect amino acids on protein antigens that are important for antibody binding.
To create the phage display polypeptide library, a random peptide sequence was displayed on the phage’s surface. Differential screening with cells as screening targets yielded polypeptides that identify specific cells. We can learn more about the receptor protein expressed specifically on the cell surface by studying the polypeptide sequence.
Protein-protein interaction studies
Almost all biological processes are mediated by protein-protein interactions. Understanding these interactions in depth is thus a major goal of modern biological chemistry. Phage display is an effective and adaptable method for studying protein-protein interactions. It can be applied to a wide range of protein interaction partners and used in a variety of applications, most notably mapping intracellular interactions of distinct protein domains.
Bacteriophage polypeptide libraries are made up of random short peptide sequences of varying lengths. Target proteins can use affinity screening to find short peptide sequences in a random library (such as receptors). We can study the interaction of two proteins after performing sequence analysis and synthesis of corresponding short peptides.
Recombinant Antibody Production
Recombinant antibodies can be used for therapeutic, diagnostic, and research purposes. As a result of the major development of molecular, numerous display technologies for recombinant antibody production have been successfully introduced. Despite this, antibody phage display is still the gold standard for producing recombinant antibodies. Its success can be attributed primarily to the phage particles’ robust nature, which enables automation and adaptation to changes.
a. Production of gene fragment
This phase entails immunizing animals with the desired antigen, followed by B lymphocyte isolation, mRNA extraction, and cDNA synthesis. The synthesized cDNA contains the genetic information for all antibodies that target different antigens and is made up of approximately 109 to 1011 lymphocyte clones.
b. Cloning of gene fragments in the phagemid vectors
Antibody-related genes are digested with restriction enzymes, cloned into phagemid vectors, and then displayed on the surface of phages. The sequence diversity of the fragments at this stage leads to optimal antibody isolation in the following steps. To package and exit from the bacterial cells and enter the medium, the phagemid vectors require the assistance of helper phage.
c. Selection of specific phages
When the fragments are cloned into phagemid vectors, a variety of antibody clones appear on the phage’s surface. Biopanning is the process of selecting specific clones that recognize the antigen (target of interest). Because of antigen-antibody binding properties, specific antibodies-carrying phages can be distinguished from non-specific phages.
d. Screening
The primary goal of this step is to isolate antibodies with high affinity for the target. Screening techniques include immunoassay, immunocytochemistry, active isolation of cells due to their fluorescent properties, and immunoblotting.
You can build your own phage display library or hire phage display companies to do it for you. You can also buy a pre-made library directly. If you already have a library, you can simply begin with the selection. Pre-made antibody libraries are a naive antibody repertoire made up of billions of different antibody scFv or fad genes. You should only use those that recognize your antigen. If the repertoire is diverse enough, antibodies against your antigen are present, and you only need to enrich them.
Protein directed evolution
As a selection-based system, phage display technology is an appealing method for the evolution of new biological drugs. Directional transformation protein refers to the use of a cassette mutation, an error-prone PCR method, or a specific cod sequence structure domain to mutant protein. Proteins or structural domain mutations found in the library will be displayed on the phage surface. Then, using affinity screening, we can obtain the required directional change of phage clones. The primary structure can be derived from a DNA sequence that can be used to screen for more cytokine receptors, new enzyme inhibitors, DNA transcription factors in combination with new sites, new cytokine antagonists, new enzymes, and to enhance biological active protein.
Drug discovery
As biologically active molecules in hormones, neurotransmitters, cytokines, antigens, and growth factors, peptides play a role in a wide range of biological processes. As a result, peptides are widely used as therapeutic and diagnostic agents in a wide range of medical fields, such as oncology, endocrinology, urology, and obstetrics. Peptide phage libraries, which contain a large number of different peptides that mimic the genuine epitopes, are useful in the development of new therapeutic peptides. So far, phage display technology has been used to develop a number of peptide drugs.
In vitro diagnostic
Currently, phage display library technology is widely used to investigate host-pathogen interactions, develop disease diagnostic markers, and identify vaccine candidates and new antibodies against infectious agents targets.
Pathogen cell-surface antigens as molecular binding sites are suitable targets for vaccine development because these antigens can affect bacterial division, replication, and virulence. There are two distinct approaches to using phage display technology in the field of infectious diseases: Pathogen-targeted phage display screens for molecular targets such as cell replication enzymes or host-pathogen virulence factors, whereas the cell-based screening method screens for bacterial whole cells. The native cell surface proteins can be targeted using cell-based screening with live pathogens. When the target antigen is unstable under immobilization conditions, this screening method is appropriate.
Read more about the procedure of phage display here

Very interesting post