 |
| plaque assay |
A streak plate for single colonies will also be prepared.
A few notes on bacteriophage:
Bacteriophages are viruses. This means that they need a suitable host cell to reproduce. The phage stock you will be titrating contains the bacteriophage lambda cloning vector CH4 with a P. ochraceus EcoRI genomic library cloned in it. The host cell used to grow the phage will be Escherichia coli strain C600 (you can use any host of your choice). Because the binding site for phage lambda is a maltose transport protein, E. coli cells used as a host for lambda must be grown in media containing maltose to induce expression of the lamB gene.
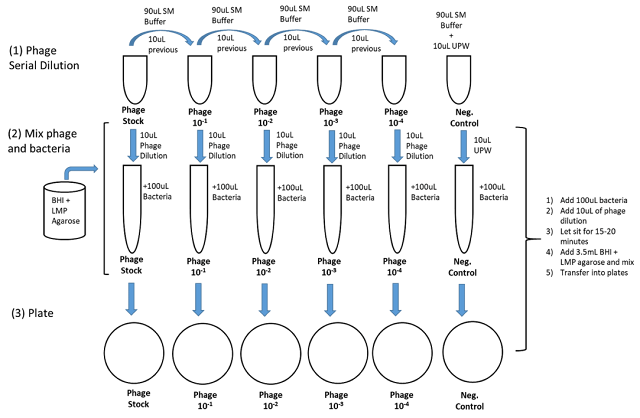
Plate cultures of bacteriophage are prepared by combining phage with susceptible host cells in top agar overlays (on top of regular nutrient agar plates). Complete agar preparations contain lower concentrations of agar (7 g/L) than standard solutions used to prepare agar plates (15 g/L) (read more about overlay technique here). The low agar concentration allows progeny phage from lysed cells to diffuse through the media and infect neighboring bacterial cells. When these cells are lysed, a plaque (zone of lysed cells) is produced on the plate. Because phage can only reproduce in actively growing cells, the plaques' size will depend on how soon the bacteria in the agar reach the stationary phase and stop copying. Plaques will stop spreading at this point.
Top agar overlays are prepared by mixing phage dilutions with susceptible bacteria and adding ~ 3 ml of liquid top agar at 45-50°C. Overlays are gently mixed and then poured on prewarmed nutrient agar plates. (Top agar must be maintained at 45-50°C until immediately before use – higher temperatures will kill bacteria, and lower temperatures will allow the agar to solidify prematurely.)
charging:
Bacteriophage particles will sometimes adhere to plastic or glass. If particles adhere to pipettor tips during transfers, the number of phage particles present in the sample transferred to the following dilution blank may be lower. One way to minimize this problem is to "charge" the pipettor by pipetting a sample of the solution up and down in the tip and discharging the sample back into the original tube. A fresh sample can then be taken from the same tube and transferred to the following dilution blank. The coating of phage particles already adhering to the walls of the tip should minimize losses in the second sample.
phage mixing:
Because top agar is a viscous solution, standard vortexing introduces air bubbles into the media, which may be difficult to distinguish from phage plaques. To reduce the number of air bubbles resulting from mixing, molten agar solutions should be mixed appropriately. The exact technique will depend on the type of tube the samples are prepared in.
A "phage mixing" technique has been developed for samples in standard glass test tubes. This is done by holding a tube with the cap in the palm of the hand and the fingers firmly holding on to the tube. The tube is held at a 45° angle and rotated in a circle 4-6 times in 1-2 seconds. (The tube must remain at 45° while being rotated – this will be demonstrated in class.) This technique allows for a gentle but thorough mixing, introducing minimal bubbles.
If the samples are prepared in 4 ml plastic tubes with snap-on caps (like they will be in this lab), the most efficient method of mixing the solutions will be to cap the tubes firmly shut and gently invert the tubes 4-6 times in 2-3 seconds. Do not mix for longer than this, as the agar will start to cool and solidify.
Phage Dilutions:
The phage aliquots provided are 1/10 dilutions (in SM) of an original phage stock with 107 and 108 pfu/ml. (pfu = plaque-forming units – these are viable phage particles capable of infecting susceptible host cells). Calculate the dilutions you will need to get final numbers of 50-500 plaques per plate. (Note that 0.100 ml of phage solution will be added to the top agar blanks – this is equivalent to an extra 1/10 dilution step.)
Dilutions will be done using SM (suspension medium). This is because phage lambda capsids require Mg2+ ions for stability. If dilutions are done in dH2O (or especially TE, which contains EDTA, a chelating agent for divalent cations), viable phage counts will be reduced.
Materials and Methods for phage titration
Materials
- 1/10 dilution (in SM) of phage stock (CH4 with a P. ochraceus EcoRI genomic library)
- 1 ml E. coli C600
- (prepared by growing an overnight culture in 10 ml LB media containing 100 microlitres filter-sterilized 20% maltose, then centrifuging and resuspending in 10 ml SM)
- SM (suspension medium)
NaCl 5.8 g
MgSO4 · 7 H2O 2 g
1 M Tris, pH 7.5 50 ml
2% (w/v) gelatine solution 5 ml
dH2O to 1 L
- 4 ml plastic Falcon tubes
- LB top agar (melted in microwave cooled to 50°C in a water bath)– do not remove from water bath until immediately before use
- disposable pipettes
- LB plates (prewarmed in 37°C incubators)
- E. coli C600 stock plate
Methods – Part I – Phage dilution
1. Obtain an adequate number of LB plates to allow 1 LB plate for every phage dilution in the range expected to produce valid plate counts. (Plates are prewarmed in 37°C incubators.) Properly label the plates, and return them to the 37°C incubators. (Labels should include group name, date, phage dilution, and media used.) (Top agar will not set as fast on prewarmed LB plates – this will allow phage overlays to be spread more evenly.)
2. Prepare serial 1/10 or 1/100 dilutions of the phage stock using 0.9 ml or 0.99 ml SM blanks. The first dilution should be 1/100 to minimize the volume of the original phage stock used. Dilutions in the range expected to produce valid plate counts should be 1/10 dilutions. (The exact number of tubes will depend on the number of dilutions you have planned in your dilution scheme.) Losses of phage can be reduced using "charging" of tips (see introduction). Dilutions must be performed aseptically.
3. Transfer 0.100 ml of each phage dilution that is to be tested to a 4 ml Falcon tube.
4. Add 0.100 ml of bacterial culture to 0.100 ml phage samples. Mix briefly, and incubate at room temperature for 15 minutes.
5. Add 3 ml top agar to tubes. Gently mix the tubes and immediately pour onto prewarmed LB plates. Spread overlay across the plate by tilting and rotating the plate until the overlay is evenly distributed. The rim of the sample tube can also be used to spread overlay and pop any bubbles present. Do not attempt to apply overlay further once it starts to set. This will produce a grainy, opaque overlay, which will make plaques challenging to see.
6. Allow plates to cool until the agar has set. Invert the plates (lid side down), and incubate 12-24 hours at 37°C. Count your leaves, record the results in your lab book, and calculate pfu concentration in the original stock.
Part II – Streaking for isolated colonies
Transfer a single colony from the E. coli C600 stock plate to an adequately labeled LB plate and streak for isolated colonies. (If you are not familiar with this technique, ask a TA for a demonstration.)
For more protocols please visit The Phage protocols


